DeepSEQreen Tutorial
A QUICK BRIEFING
DeepSEQreen offers three distinct functionalities tailored to meet the specific needs of users:
- Drug hit screening
Designed for users who aim to identify potential drug hits within a chemical library for their target protein of interest.
- Target protein identification
Intended for users who possess a specific drug or compound and seek to discover potential target proteins associated with it.
- Interaction pair inference
Geared towards users with paired targets-and-compounds, enabling them to determine the interaction strength between these pairs. Additionally, it allows users to explore interactions and strengths among various combinations of target-compound pairs when provided with separate lists of target proteins and compounds.
This illustrated tutorial aims to provide a step-by-step instruction on how to effectively utilize each of these functionalities.
Launch DeepSEQreen
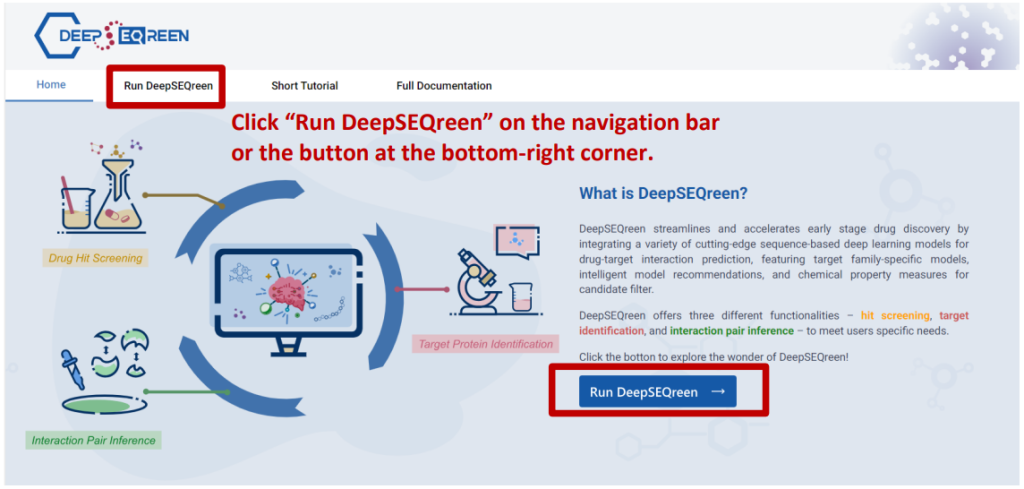
Drug Hit Screening
There are five major steps before you can submit your job to search for potential drug hits for your designated target protein:
Step 1: Target protein input
We accept three forms of input: UniProt ID, gene symbol and sequence in FASTA format.
Option 1: input a protein sequence
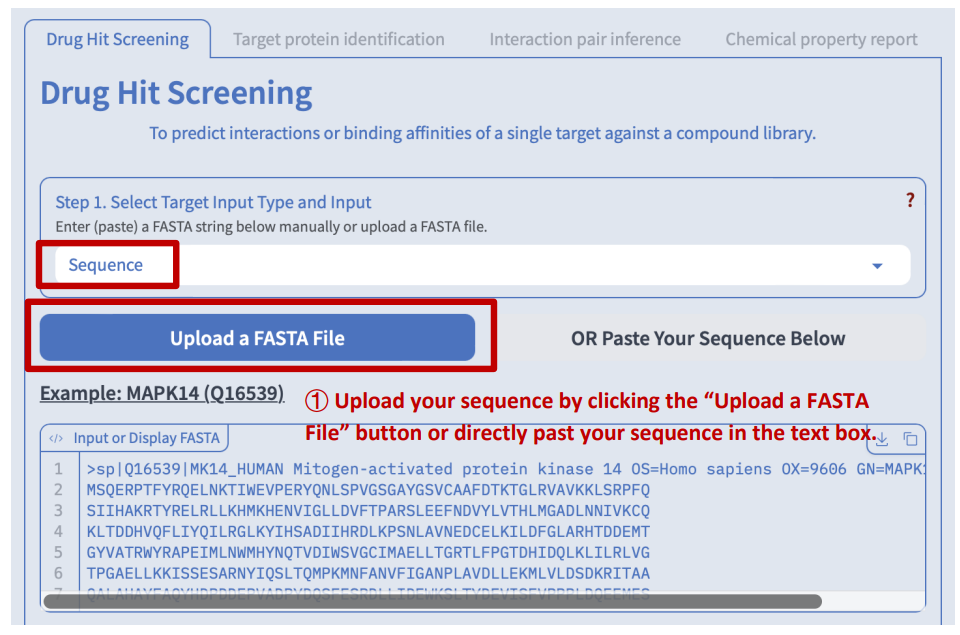
Hint: Hover the mouse over the question mark in the upper right corner of each step to view a brief introduction and instructions for that particular step.

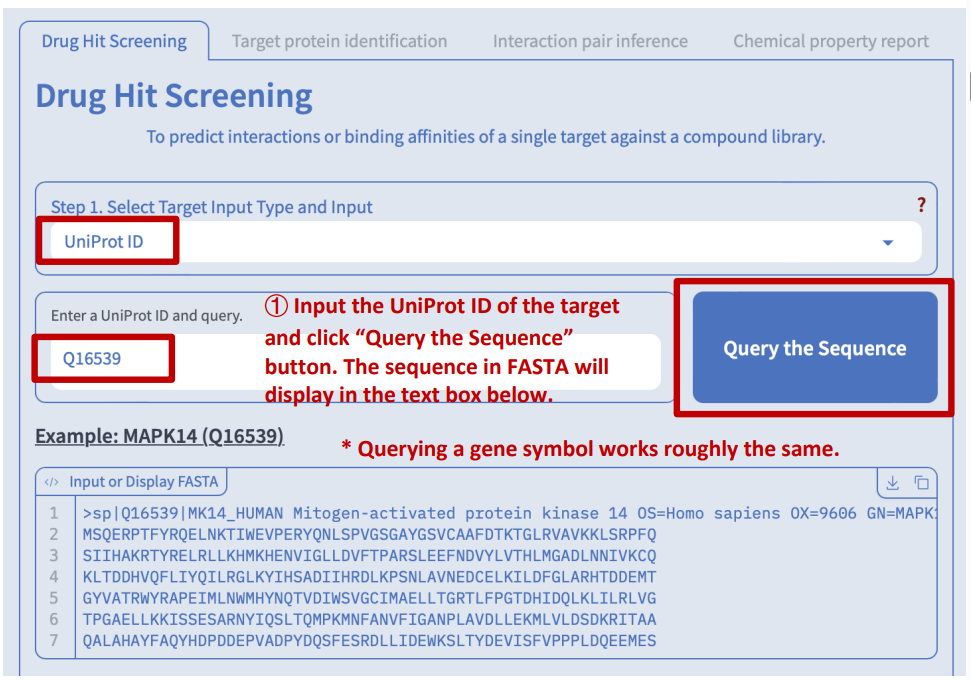
Option 2 and 3: input an UniProt ID or gene symbol
Step 2: Select the target family
DeepSEQreen offers target family-specific models to enable precise prediction of potential drug hits for user-submitted target proteins. Users can select protein types from various families, including kinase, non-kinase enzyme, membrane receptor, nuclear receptor, ion channel, other protein targets, general (family non-specific), etc. Alternatively, they can utilize the auto-detection feature which suggests the most likely family type based on BLAST results. This allows for tailored and accurate predictions based on the specific protein of interest. (Hint: If the target family is not specified, DeepSEQreen will utilize the general model for prediction.)
Step 3: Select or upload a compound library
DeepSEQreen currently provides two pre-set compound libraries: DrugBank and Drug Repurposing Hub. More pre-set libraries will be added soon. Additionally, users have the flexibility to upload their own custom library in either SDF or CSV format. Example files are available for users to try out the functionality or download in order to guide them on the required formatting for their custom libraries. Users can also refer to the DeepSEQreen documentation for detailed instructions on the formatting requirements for custom libraries.
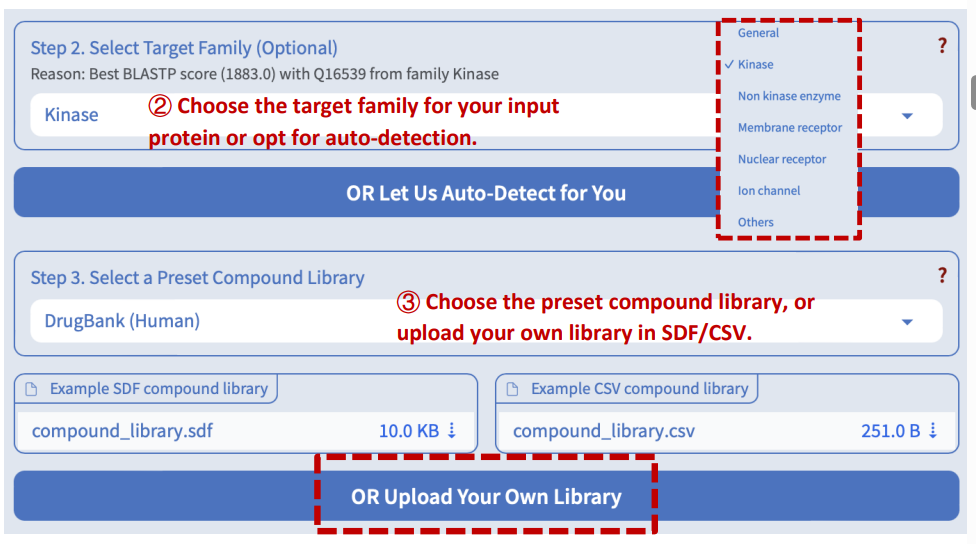
Step 4: Select the prediction task
DeepSEQreen offers two prediction options: 1) the probability of the interaction between the input protein and the compounds in the library, and 2) the binding affinity between them.
Step 5: Select a preset model
DeepSEQreen offers ten preset models. Users can select any of them or choose auto-detection, which recommends the model with the highest AUROC (for interaction task) or CI (for binding affinity task).
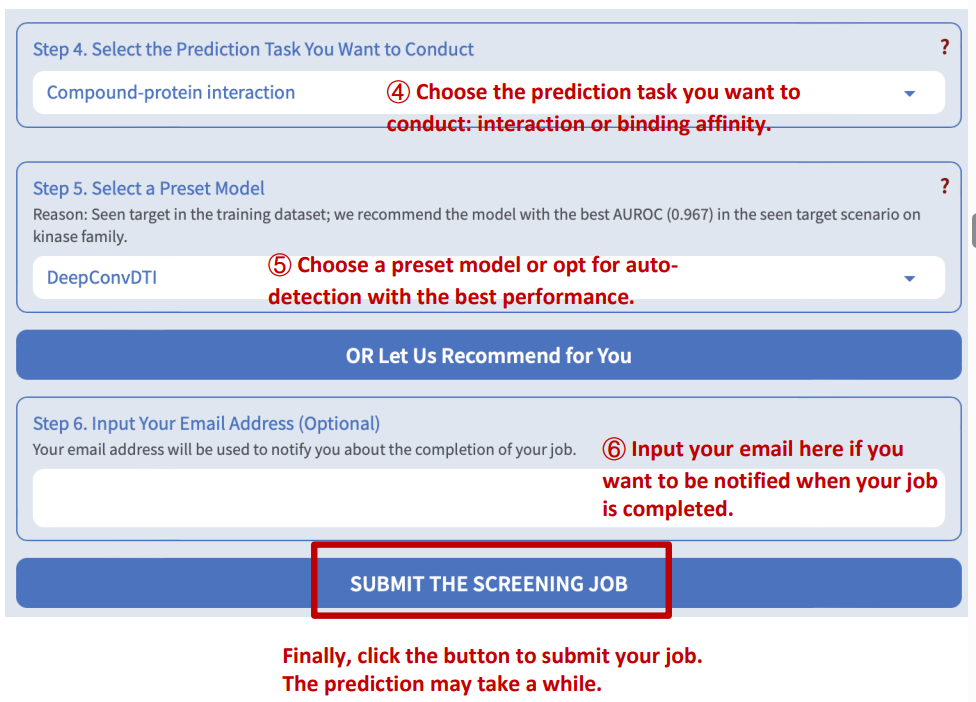
Results will automatically display when the screening job is completed.
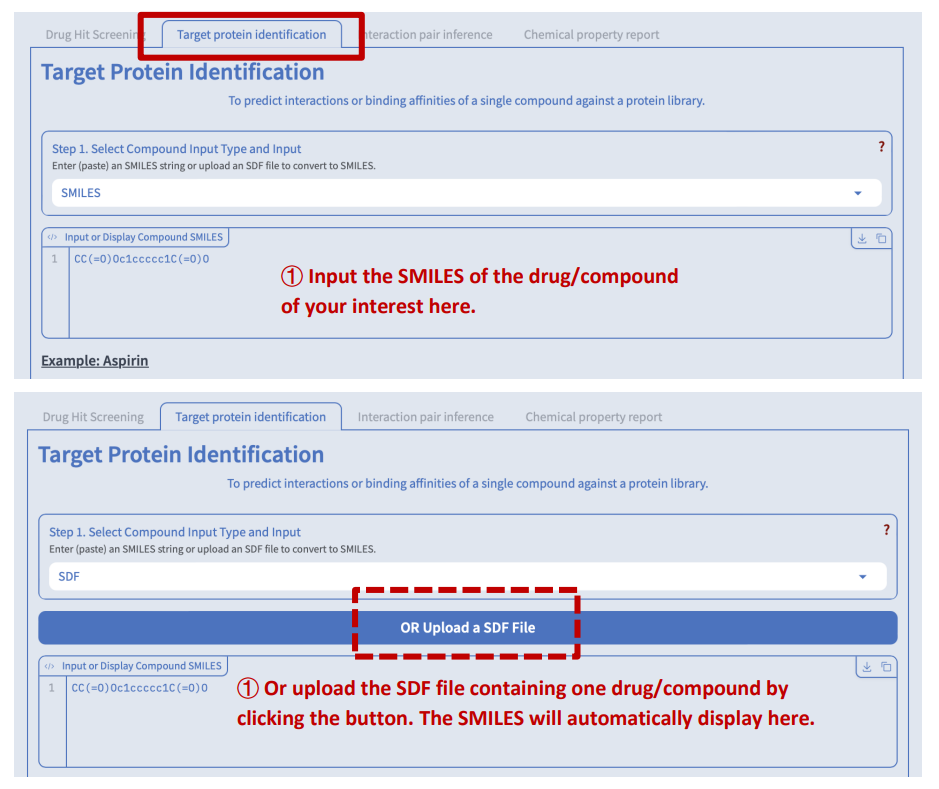
Target Protein Identification
Switch to the “Target protein identification” tab. There are also five major steps before you can submit your job to search for possible target proteins for your drug or compound:
Step 1: Compound input
We accept two forms of input: SMILES and SDF.
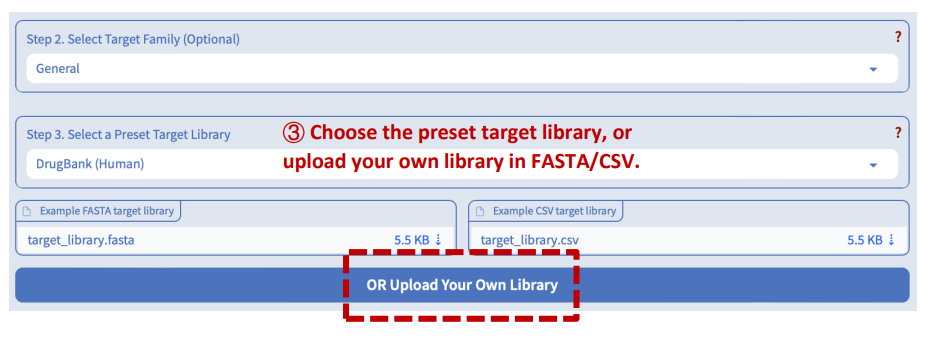
Step 3: Select or upload a protein library
DeepSEQreen currently provides two pre-set protein libraries: DrugBank and ChEMBL (v33). More pre-set libraries will be added soon. Additionally, users have the flexibility to upload their own custom library in either FASTA or CSV format. Example files are available for users to try out the functionality or download in order to guide them on the required formatting for their custom libraries. Users can also refer to the DeepSEQreen documentation for detailed instructions on the formatting requirements for custom libraries.
Step 4: Select the prediction task
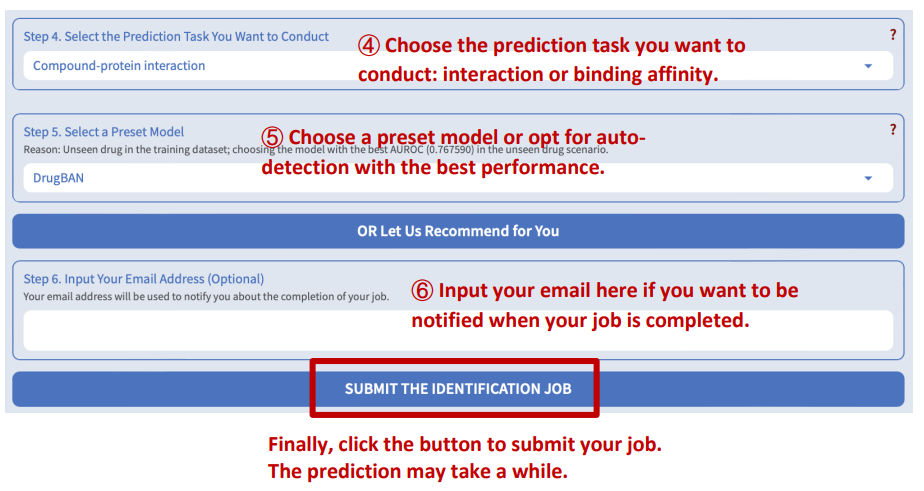
DeepSEQreen offers two prediction options: 1) the probability of the interaction between the input protein and the compounds in the library, and 2) the binding affinity between them.
Step 5: Select a preset model
DeepSEQreen offers ten preset models. Users can select any of them or choose auto-detection, which recommends the model with the highest AUROC (for interaction task) or CI (for binding affinity task).
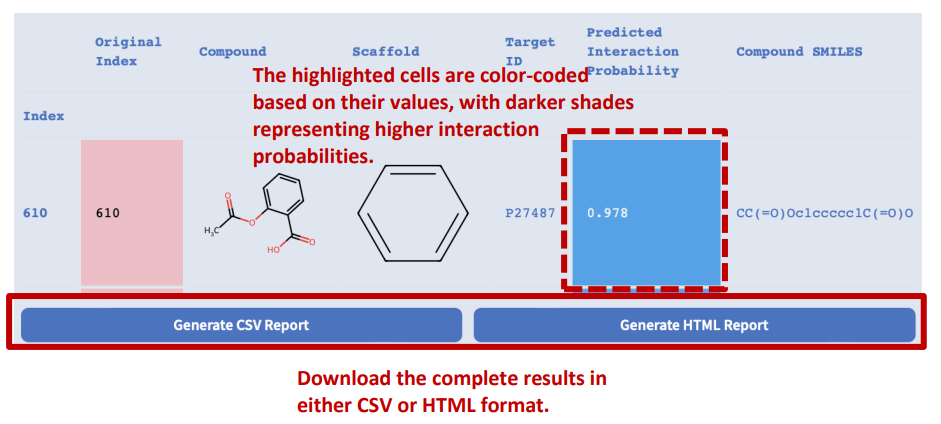
Results will automatically display when the screening job is completed.
Interaction Pair Inference
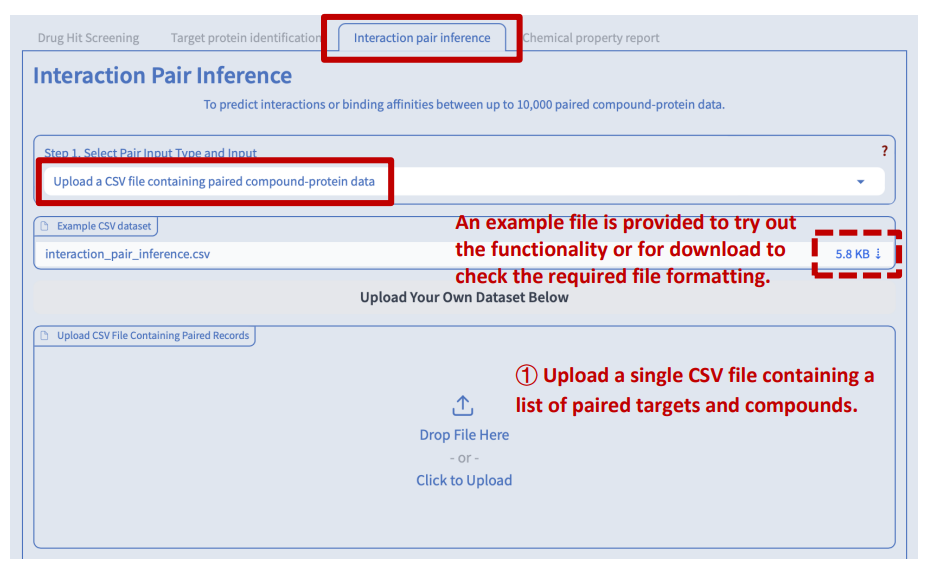
Switch to the “Interaction pair inference” tab. There are four major steps before you can submit your job to prediction the interaction probability or binding affinity between the input compounds and target proteins:
Step 1: Data input
We accept two types of input: 1) a single file containing a list of paired targets and compounds, and 2) two separate files containing lists of target proteins and compounds respectively.
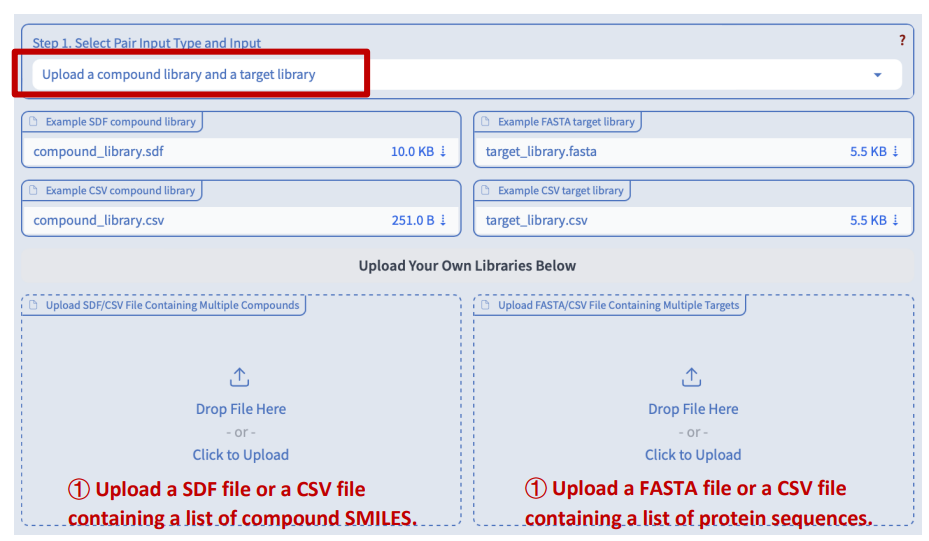
Step 2: Select target family
By default, DeepSEQreen applies models trained on all protein families (general). However, if the proteins in the target library of interest belong to the same protein family, users have the option to manually select that specific family for more accurate predictions.
Step 4: Select the prediction task
DeepSEQreen offers two prediction options: 1) the probability of the interaction between the input protein and the compounds in the library, and 2) the binding affinity between them.
Step 5: Select a preset model
DeepSEQreen offers ten preset models. Users can select any of them.
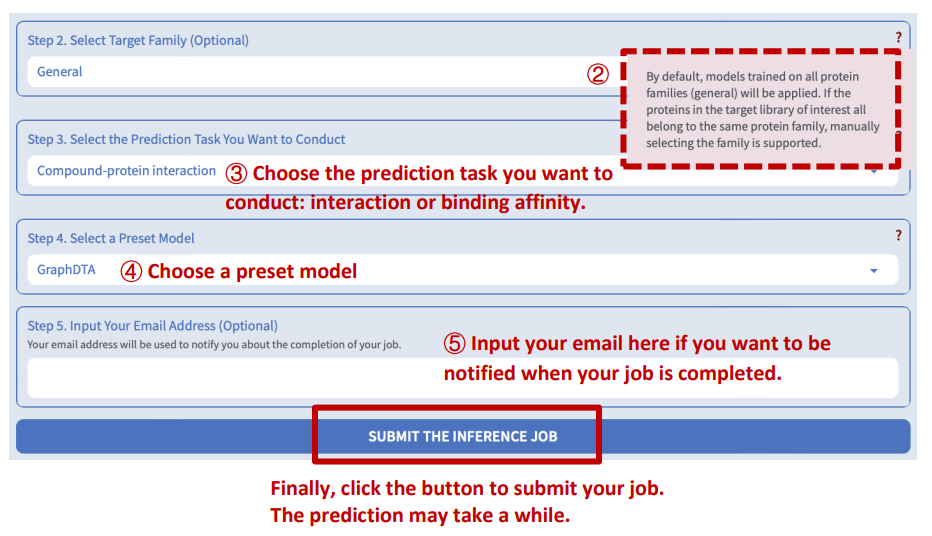
Results will automatically display when the screening job is completed.
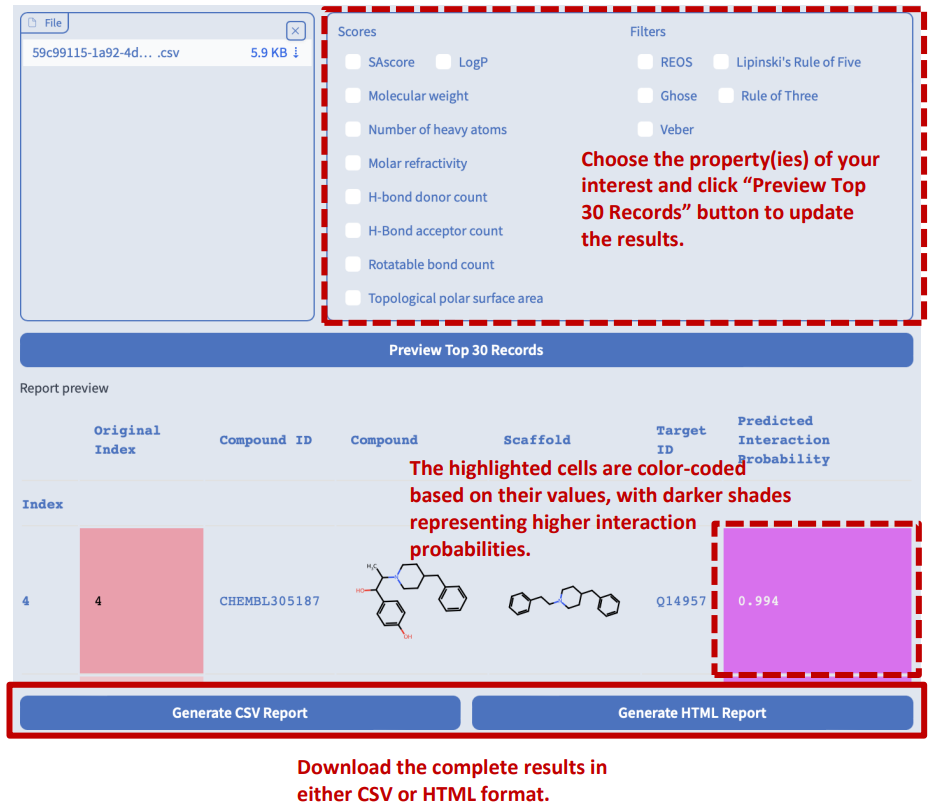
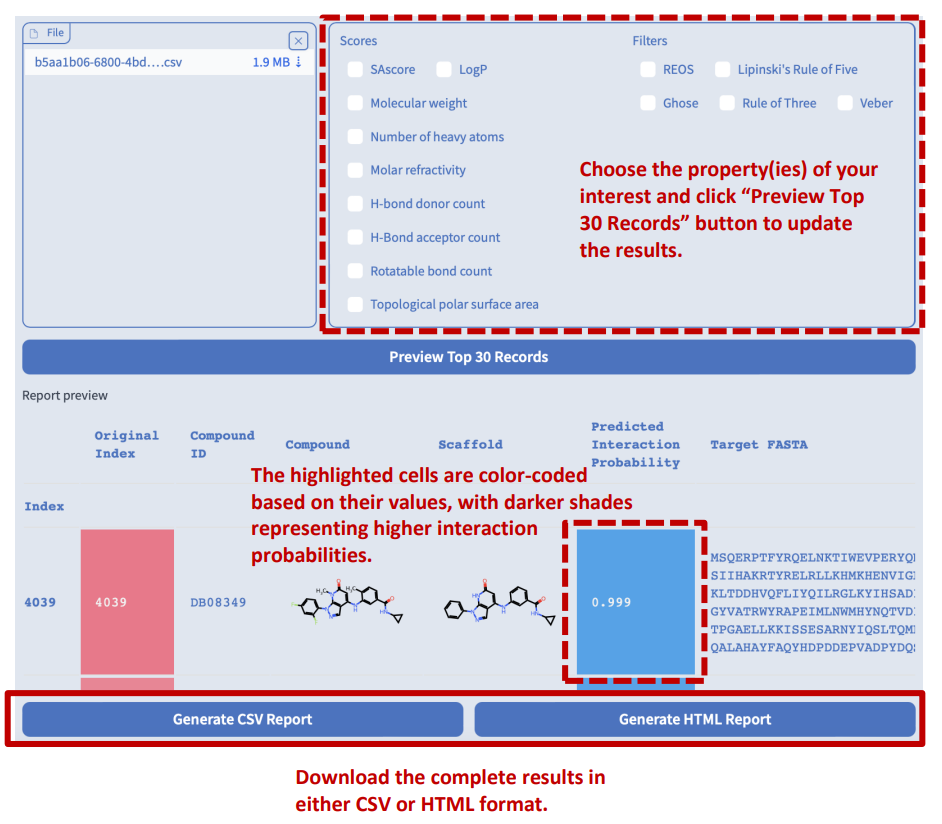
Chemical Property Report
Switch to the “Chemical property report” tab. In addition to displaying preview reports for drug hit screening, target protein identification, and interaction pair inference tasks, users can also upload a CSV file containing a list of compound SMILES to calculate their basic chemical properties, synthetic accessibility measures, and drug-likeness.
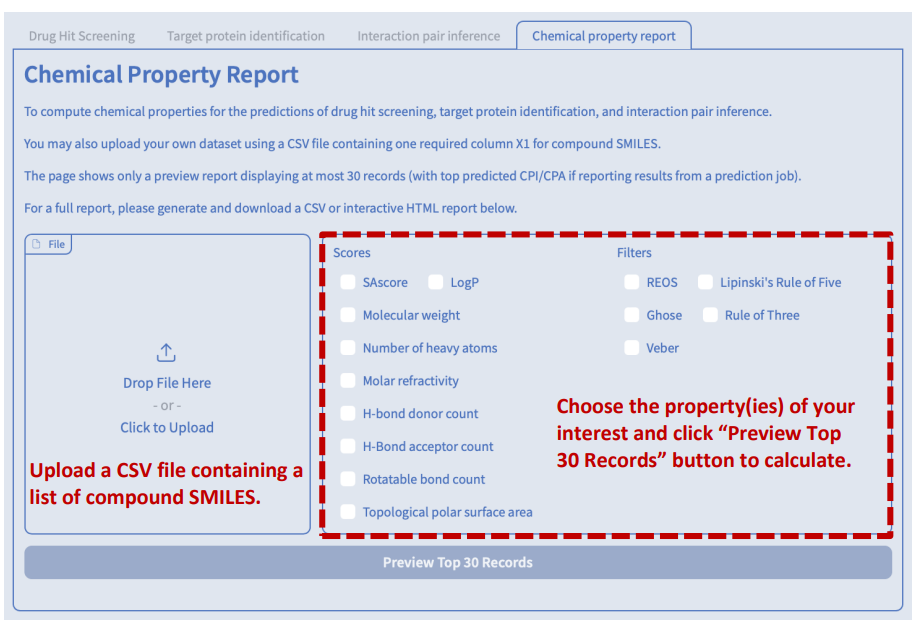
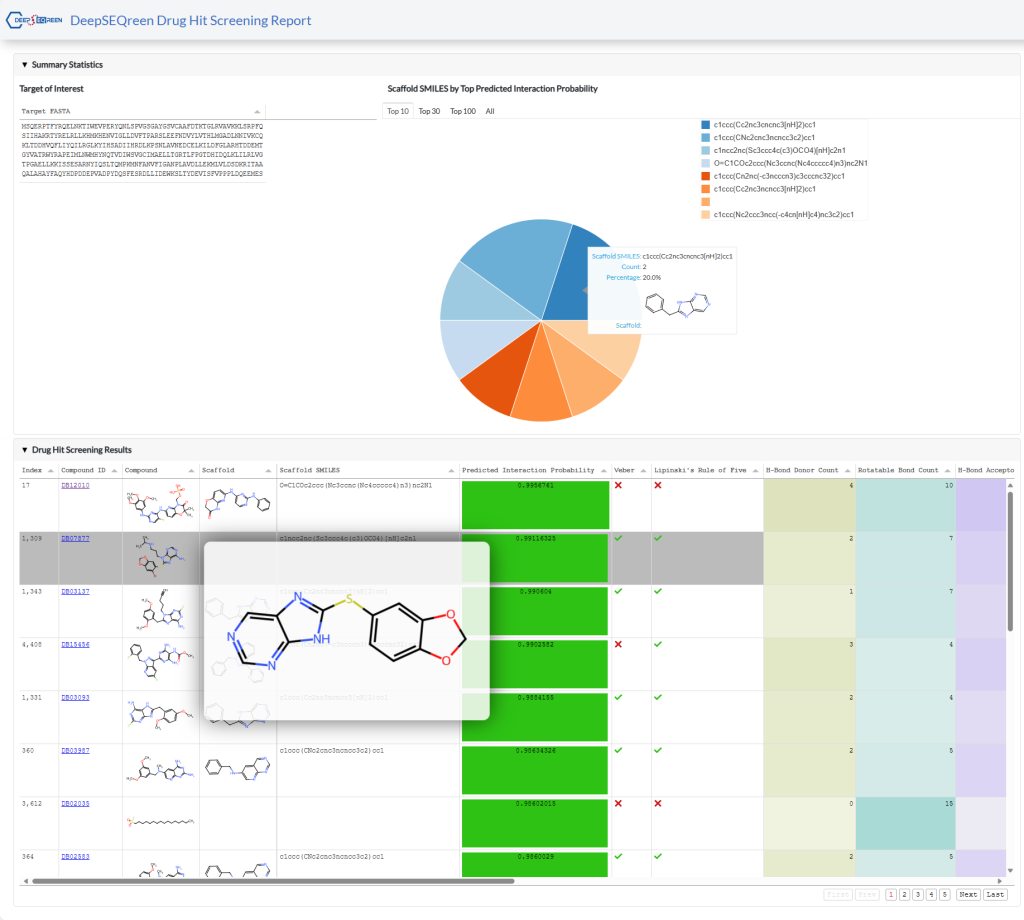
This is an example of an interactive HTML report generated by Drug Hit Screening. You can explore the report in the following ways:
- Hover your cursor over some elements to see tooltips with more information or enlarge images.
- Click on the column header of any table to sort the data by that column.
- Click on the links in the Compound ID or Target ID column to view the details of the compound or target in the UniProt or PubChem databases.