About GenFBDD
FBDD traditionally depends on experience-demanding and intuition-driven workflows, including but not limited to fragment docking, hotspot identification, and linker design. GenFBDD is a pipeline and web tool that reimagines the FBDD process with a three-stage Dock-Link-ReDock pipeline, empowered by SOTA molecular generation models like DiffDock and DiffLinker, to mirror, streamline and automate FBDD.
Getting Started
GenFBDD requires only two inputs: a set of chemical fragments and a target protein structure.
Chemical Fragment Library
You can choose from a list of fragment libraries or upload your own fragments to GenFBDD. Uploading a custom fragment library supports two file formats:
– SDF: a standard Structure Data Format file that stores the molecular data of multiple fragments.
– CSV: a comma-separated values file that MUST contain the following columns:
– X1: the SMILES strings of the fragments.
– ID1: the IDs of the fragments. Note that each SMILES string must have a unique ID.
When you select or successfully upload a fragment library, GenFBDD will display a tabular preview of the fragments.
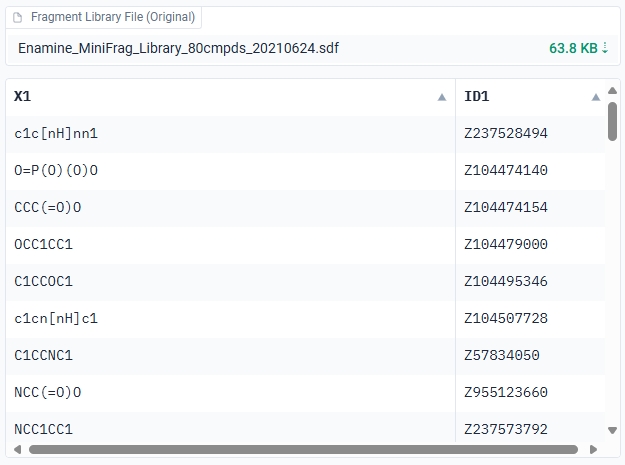
Preprocess the Fragment Library
GenFBDD will automatically preprocess the fragment library by complete fragmentation of unconnected molecules and removal of duplicates. Moreover, there are several custom preprocessing options available:
– Dehalogenate Fragments: Replaces halogen atoms with implicit hydrogen atoms from the fragments.
This is useful if you want to generate more scaffold-like lead compounds.
– Remove Inorganic Fragments: Removes metallic or inorganic fragments from the library.
After clicking on Process Fragments, the changes will be applied and displayed on the tabular preview as well.
Target Protein Structure
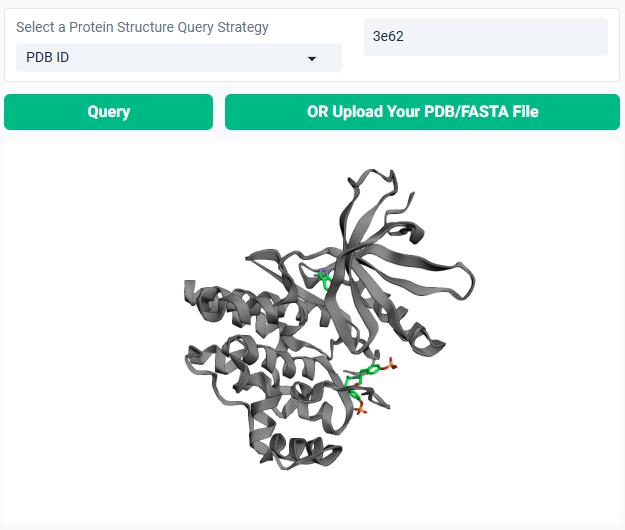
You can query the PDB database on GenFBDD with a PDB ID, a UniProt ID, or a sequence of the protein. GenFBDD will download the PDB file of the most relevant query result. You can also upload your own PDB file.
When you successfully query or upload a protein structure, GenFBDD will display a 3Dmol preview of the protein structure. You can interact with the 3D preview in similar ways with molecular viewers like PyMol or Jmol.
| Movement | Mouse Input | Touch Input |
|---|---|---|
| Rotation | Primary Mouse Button | Single touch |
| Translation | Middle Mouse Button or Ctrl+Primary | Triple touch |
| Zoom | Scroll Wheel or Second Mouse Button or Shift+Primary | Pinch (double touch) |
| Slab | Ctrl+Second | Not Available |
Extract Pocket from the Protein
GenFBDD provides three approaches to extract the pocket of the protein structure:
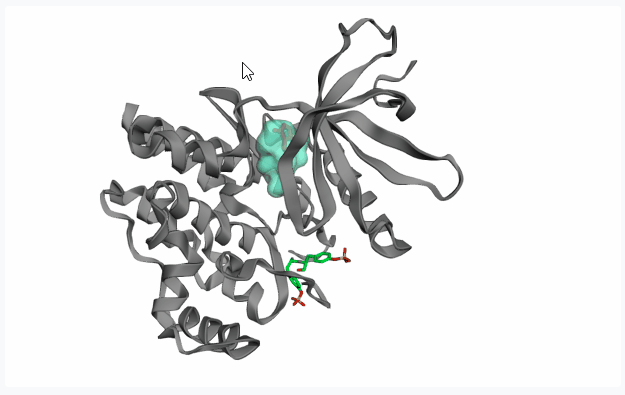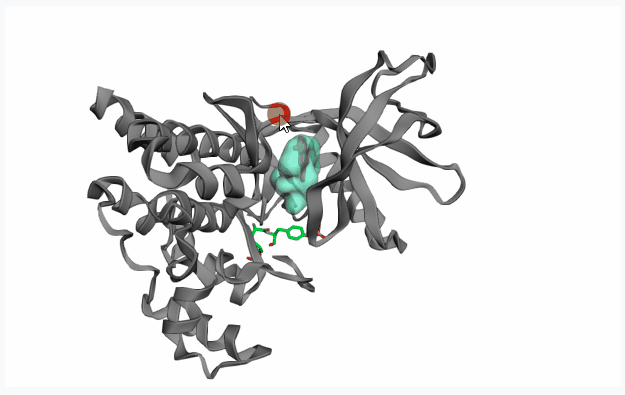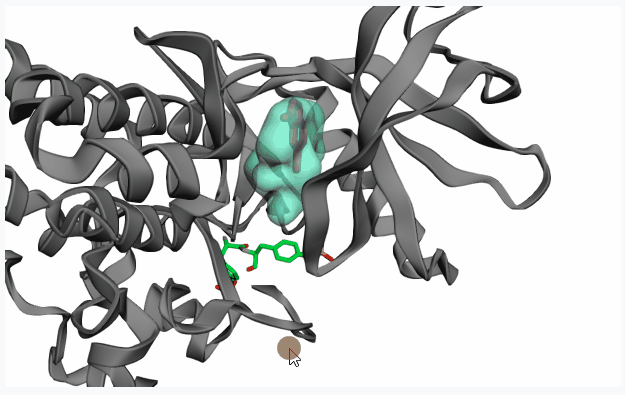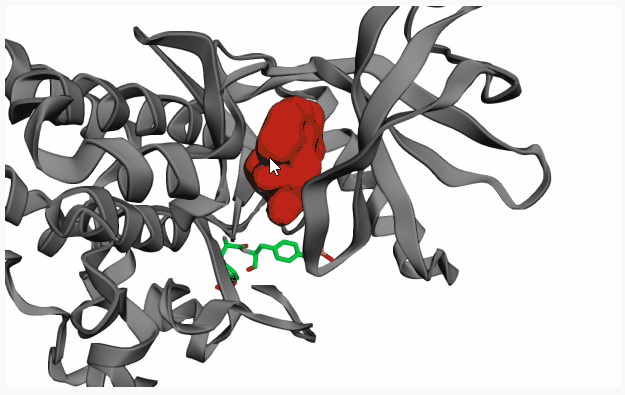
- Topological Prediction with Fpocket: Fpocket is a very efficient tool for accurate pocket detection based on topological features of the protein structure. Select this option and click on
Extract Pocketto display all pockets detected by Fpocket on the 3D protein structure preview.If your PDB structure contains a co-crystallized ligand in your pocket of interest, on the 3D preview you can click on the ligand (highlighting it in red) before clicking on “Extract Pocket” to extract only the pocket of interest.
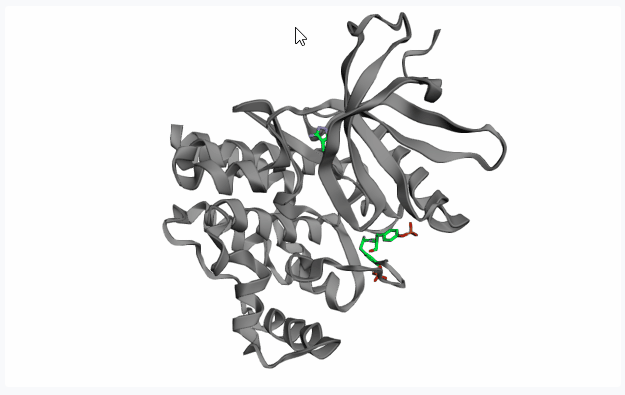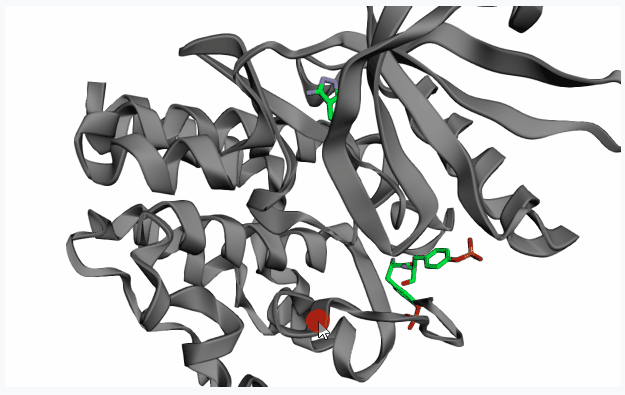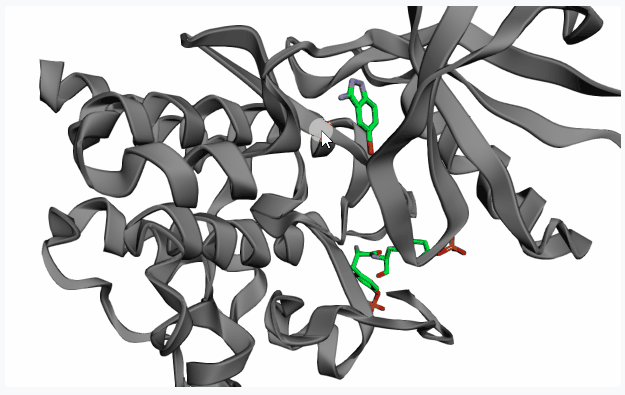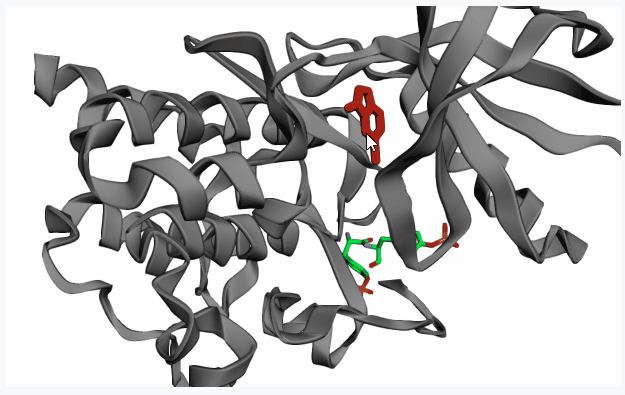
After extracting the pocket(s), please click on your pocket of interest (highlighting it in red) to select it for fragment linking.
-
Fragment Pose Clustering: This option spatially clusters the poses of the docked fragments and identifies “hotspots” where docked fragment conformers are most concentrated and treat top N clusters as potential pockets. Note that this is performed after the Dock phase, thu unable to show any pocket preview.
Dock Phase Settings
Here, you can adjust the fragment docking settings for the first stage of the pipeline.
- Number of Poses Per Fragment: The number of docking poses or conformers to generate for each fragment.
- Confidence Cutoff For Poses: The minimum confidence score for a pose to be considered for linker generation.
- Advanced Options: For users familiar with diffusion-based models.
- Number of Denoising Steps: The number of denoising steps for sampling docking poses.
Link Phase Settings
Here, you can adjust the fragment linking settings for the second stage of the pipeline.
– Allow Fragment Merging by Geometry: Allow merging a pair of fragment conformers when the distance (Å) between their closest atoms is unsuitable for fragment linking due to overlap of covalent radii.
– Fragment Pose Distance Range: The maximum distance (Å) between the closest atoms of a pair of fragment conformers to qualify them for linker generation.
– Number of Linkers Per Pair: The number of linkers to generate for each selected pair of fragment poses.
– Advanced Options: For users familiar with diffusion-based models.
– Linker Size: The size (number of heavy atoms) of the linkers to generate, where 0 means predicted, and greater or equal to 1 means a fixed size.
– Number of Denoising Steps: The number of denoising steps for sampling fragment linkers.
Results
The Results tab allows you to visually preview the generated molecules, calculate compound properties and filters, and download the result files for further analysis.
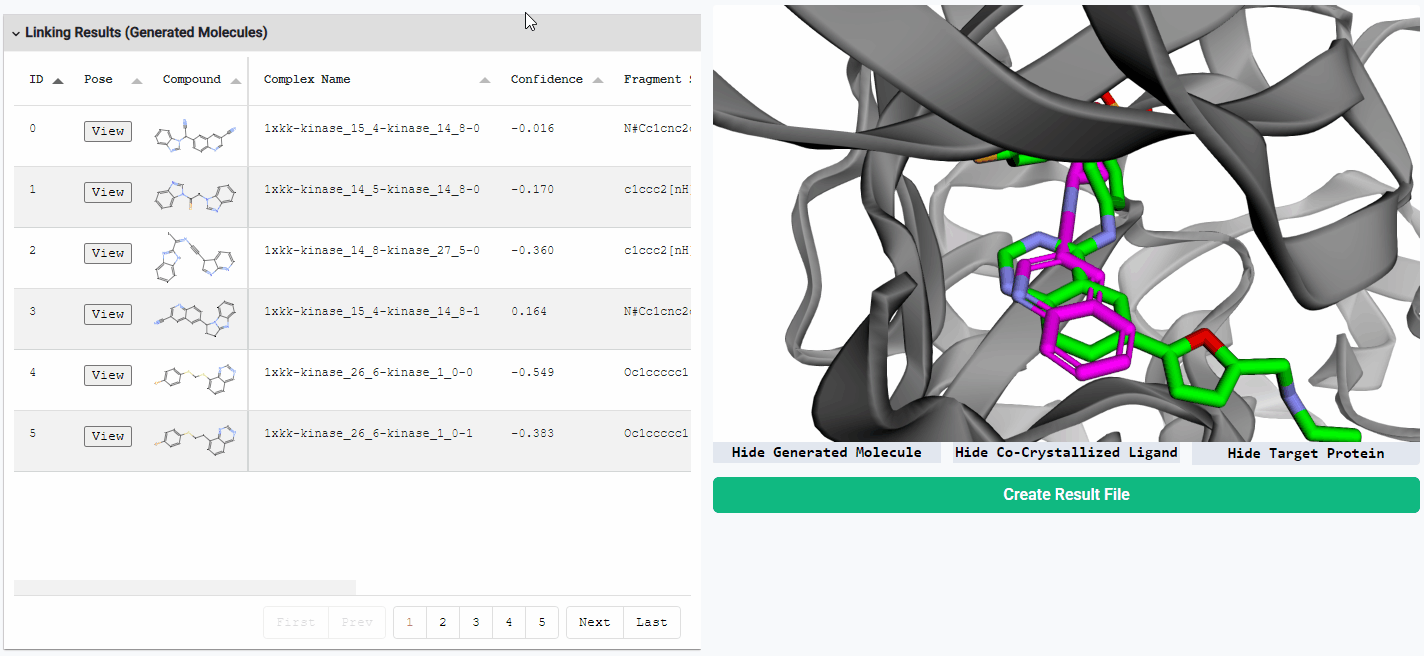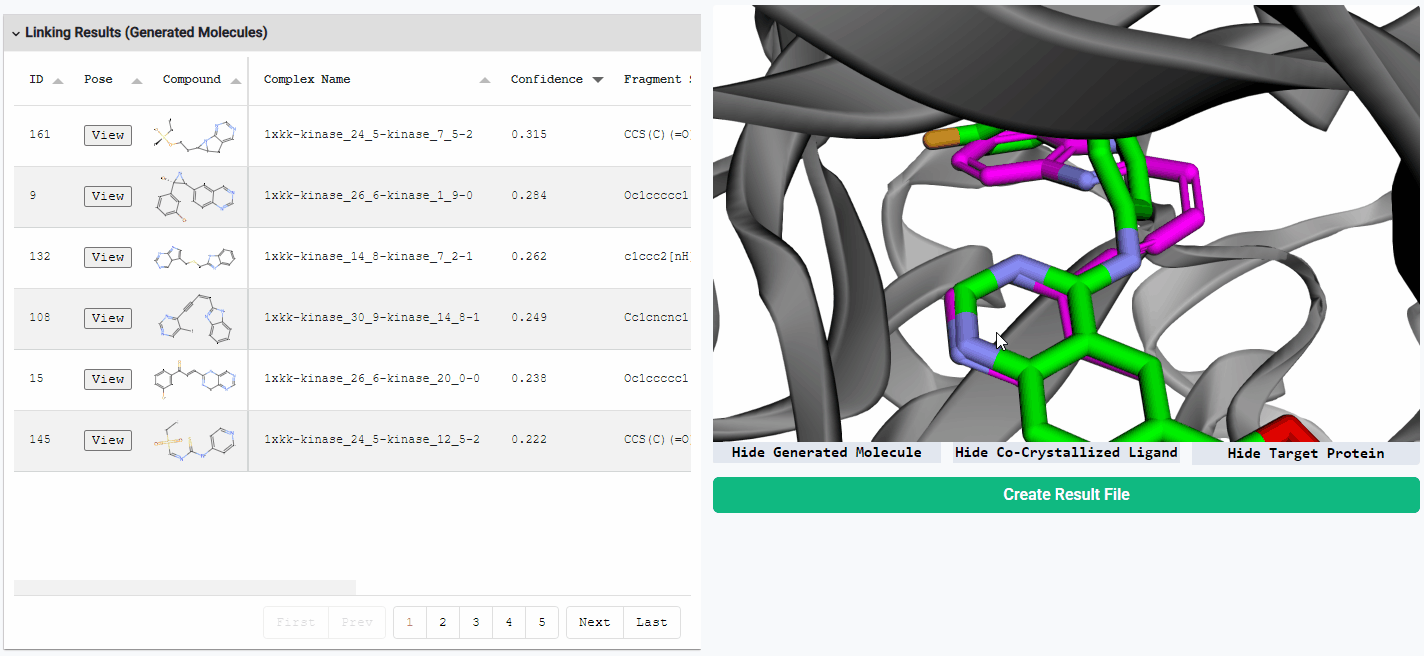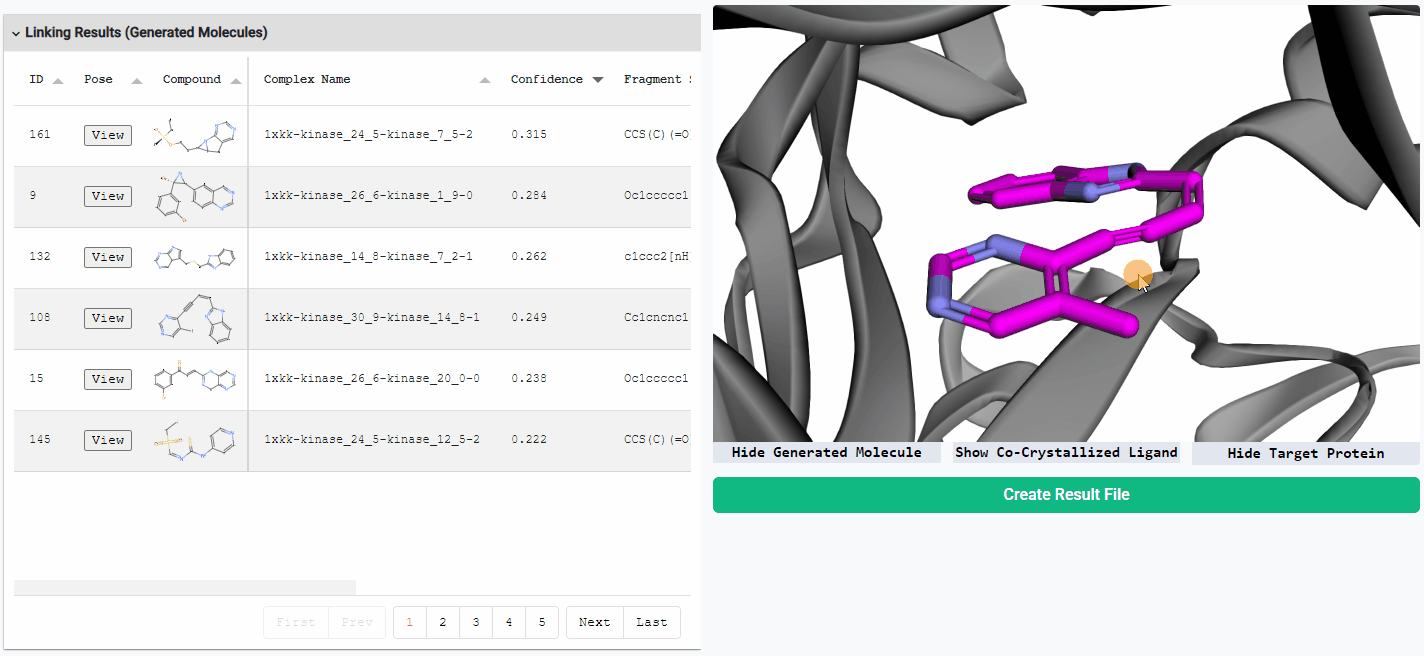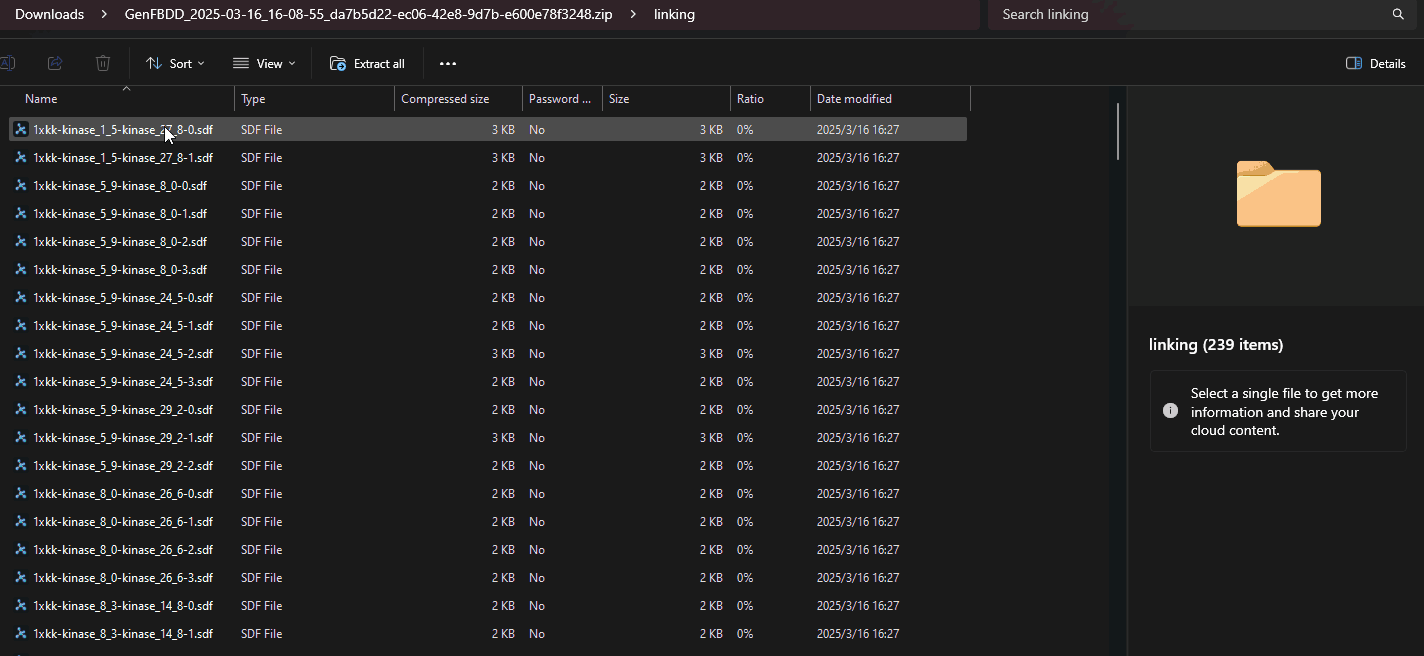
Preview Generated Molecules
To view the generated molecules in their docked conformation alongside the protein structure, simply click the “View” button in the corresponding row of the result table. You can sort the molecules in the result table by docking confidence or any other calculated properties. You can freely hide or show the generated molecule, the protein structure, and the co-crystallized ligand (if available) using the toggle buttons located below the visualization window.
Calculate Compound Properties and Filters
You can also calculate additional compound properties and apply filters to all generated molecules. Simply select your desired properties or filters and click on Calculate Properties. These calculated properties and filters will automatically appear in the preview table and will be included in the final downloadable result files.
Download Result Files
After you have calculated the desired molecular properties and filters, click on Create Result File to create a downloadable zip file containing the summary table, the protein structure, and all generated molecules. Click on ↓ next to the file name to download.